Avian Influenza Virus
Genomes
Influenza A viruses (including avian influenza) are single stranded RNA viruses, with a segmented genome. There are 8 segments encoding for 10-12 proteins, and the segments range in size from 0.9kb to 2.4kb. See for example http://viralzone.expasy.org/all_by_protein/6.html
Complete genomes are available for thousands of strains, from the Influenza Virus Resource (NCBI) (open) and GISAID (requires login).
Subtypes
Influenza viruses are categorised by the subtype of the Hemagglutinin (HA) and Neuraminidase (NA) surface proteins, for example H5N1 (HA type 5, NA type 1).
There are 17 HA types and 10 NA types.
HA 1-16 and NA 1-9 have been isolated from avian hosts (and H17N10 was found in Bats in 2012 doi: 10.1073/pnas.1116200109).
For a general introduction to influenza viruses see Webster 1992.
Avian influenza viruses circulating in the wild bird reservoir are usually low-pathogenic, however the inclusion of a cleavage site between the HA1 and HA2 subdomains within HA (typically a multi-basic cleavage site) is highly pathogenic particularly to chickens, and Highly pathogenic avian influenza (HPAI) is defined according to the effect on chickens and the presence of the cleavage site. General information on avian influenza can be found at Flu-Lab-Net : http://www.flu-lab-net.eu/about_AI.html
Currently (2014), only H5 and H7 subtypes have been known to aquire the cleavage site that results in high pathogenicity, and HPAI outbreaks have occurred in Asia, Europe, Africa and North America with subtypes including H5N1, H5N2, H5N3, H7N1, H7N3, H7N7 (not in order).
Sequences
Global sample data sets
All avian influenza complete genomes were downloaded from the Influenza Virus Resource (interface to GenBank) on 25th Oct 2013.
A sample of one per category was randomly chosen, the categories were:
- subtype
- bird type (wild/domestic + bird order, e.g. galliformes/anseriformes)
- country or state (for USA, Canada, China, Russia, UK)
- year
H5N1 in Europe, Asia and Africa
H5N1 isolates from Europe, Asia and Africa from 1996 to present were selected from the global sample of 2158 genomes (above),
resulting in a set of 238 isolates, of which the majority were HPAI (233 of 238).
Zipped fasta sequences, trees and information files of Table 1
Table 1: Sequences, Neighbour joining Trees, and sequence information from 238 H5N1 isolates from Europe, Asia and Africa
| Segment | Protein | Fasta Sequences | Newick NJ Tree | Information Table |
| 1 | PB2 | Seg 1 Sequences | Seg 1 Tree | Seg 1 Table |
| 2 | PB1 | Seg 2 Sequences | Seg 2 Tree | Seg 2 Table |
| 3 | PA | Seg 3 Sequences | Seg 3 Tree | Seg 3 Table |
| 4 | HA | Seg 4 Sequences | Seg 4 Tree | Seg 4 Table |
| 5 | NP | Seg 5 Sequences | Seg 5 Tree | Seg 5 Table |
| 6 | NA | Seg 6 Sequences | Seg 6 Tree | Seg 6 Table |
| 7 | M1 & M2 | Seg 7 Sequences | Seg 7 Tree | Seg 7 Table |
| 8 | NS1 & NS2 | Seg 8 Sequences | Seg 8 Tree | Seg 8 Table |
Figure 1: Neighbour joining tree of 238 H5N1 HA (segment 4) sequences
Grey = LPAI, Blue = European HPAI, Orange = Africian HPAI, Black = Asian HPAI.
Click to Zoom
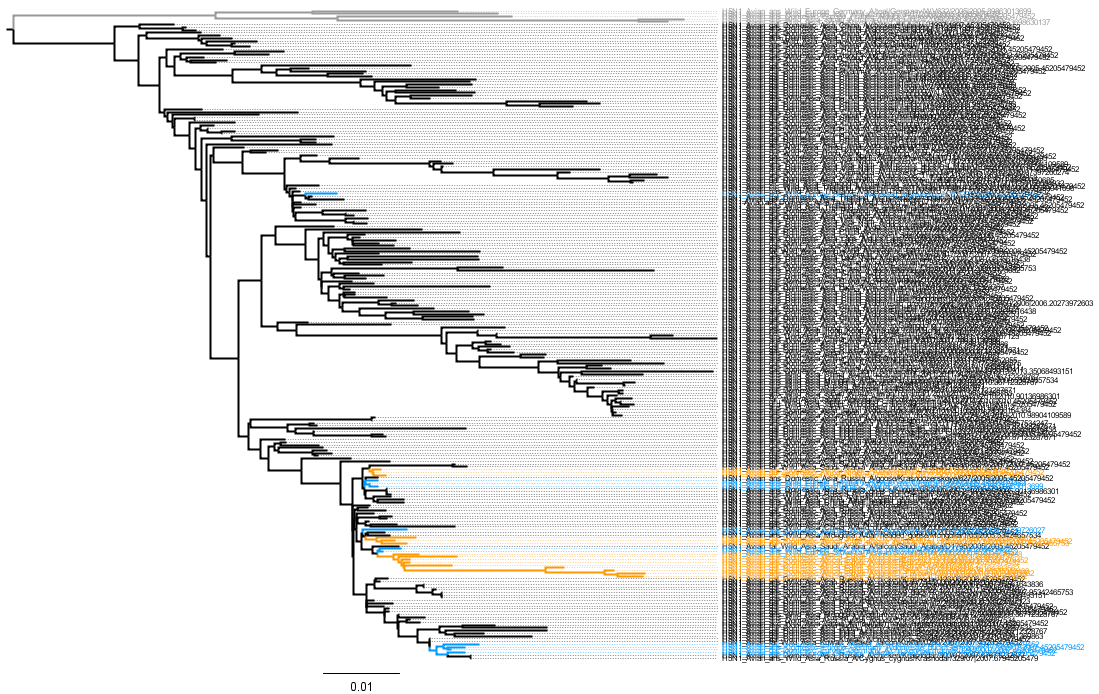
Seg 4 FigTree File
LPAI data sets from Eurasia
A study of reassortments among avian influenza viruses in Eurasia was performed by Lu et al (2014), and the data (in xml format) is available via Dryad:
Lu L, Lycett SJ, Leigh Brown AJ (2014)
"Reassortment patterns of avian influenza virus internal segments among different subtypes"
BMC Evolutionary Biology 14:16. doi:10.1186/1471-2148-14-16 -
Data on Dryad
The recently emerged H7N9 strain, which although LPAI still infected and caused death in humans in China, was sequenced and studied by Lam et al:
Lam et al (2013) "The genesis and source of the H7N9 influenza viruses causing human infections in China"
Nature 502:241–244
doi:10.1038/nature12515
Background sequences from internal protein coding segments for this study, and ongoing studies into LPAI in China: zipped nexus files
Additional studies
A study of worldwide avian H7 and its interaction with NA
Ward MJ, Lycett SJ, Avila D, Bollback J, Leigh Brown AJ (2013)
"Evolutionary interactions between haemagglutinin and neuraminidase in avian influenza"
BMC Evol Biol 2013 13:222 doi:10.1186/1471-2148-13-222
Reassortments in Eurasian swine, also contains large avian, swine and human trees (in supplementary)
Lycett SJ et al (2012)
"Estimating reassortment rates in co-circulating Eurasian swine influenza viruses"
J Gen Virol 9311:2326-2336 doi:10.1099/vir.0.044503-0
Very large scale trees from avian, human, swine and equine influenza, showing an H7 equine strain is ancestral to the modern strains
Worobey M, Han G-Z, Rambaut A (2014)
"A synchronized global sweep of the internal genes of modern avian influenza virus"
Nature 508:254-257 doi:10.1038/nature13016
